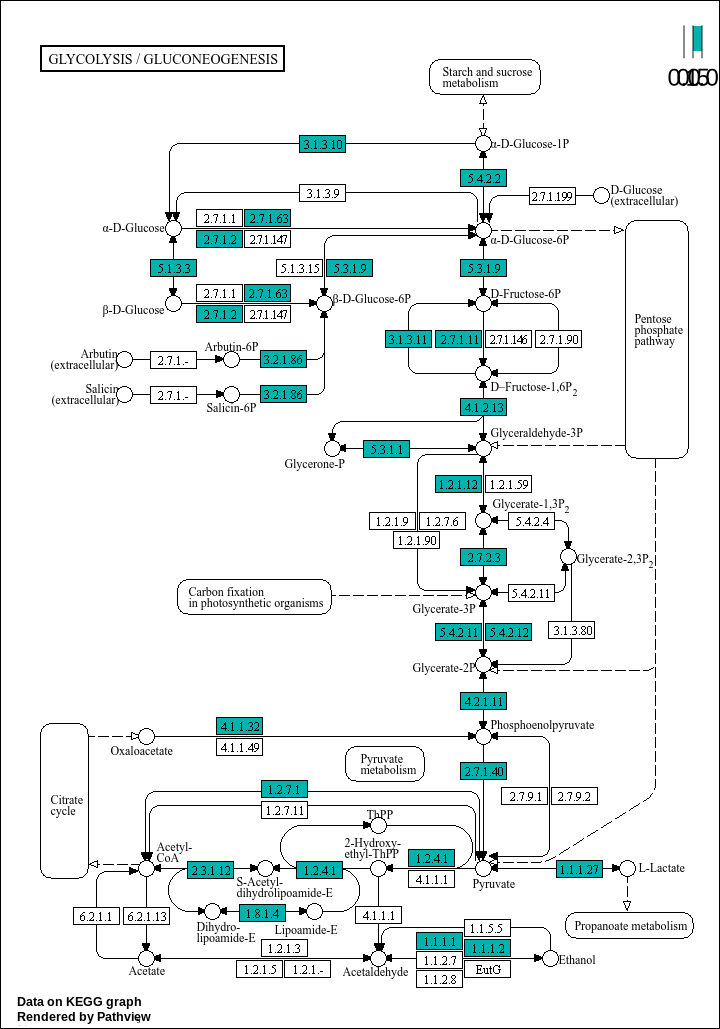
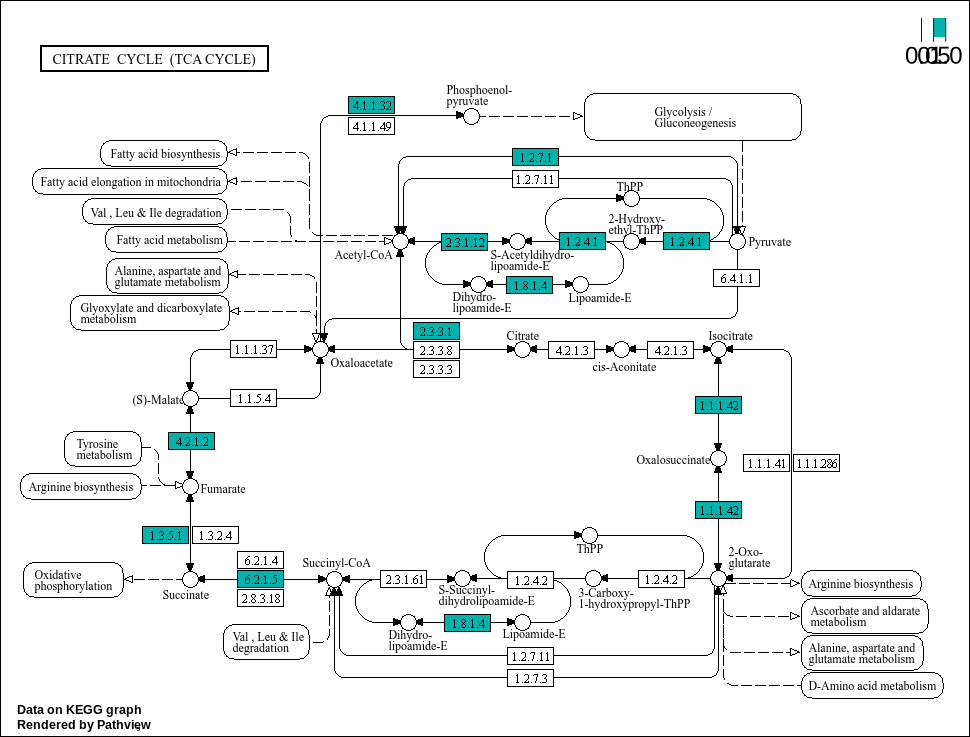
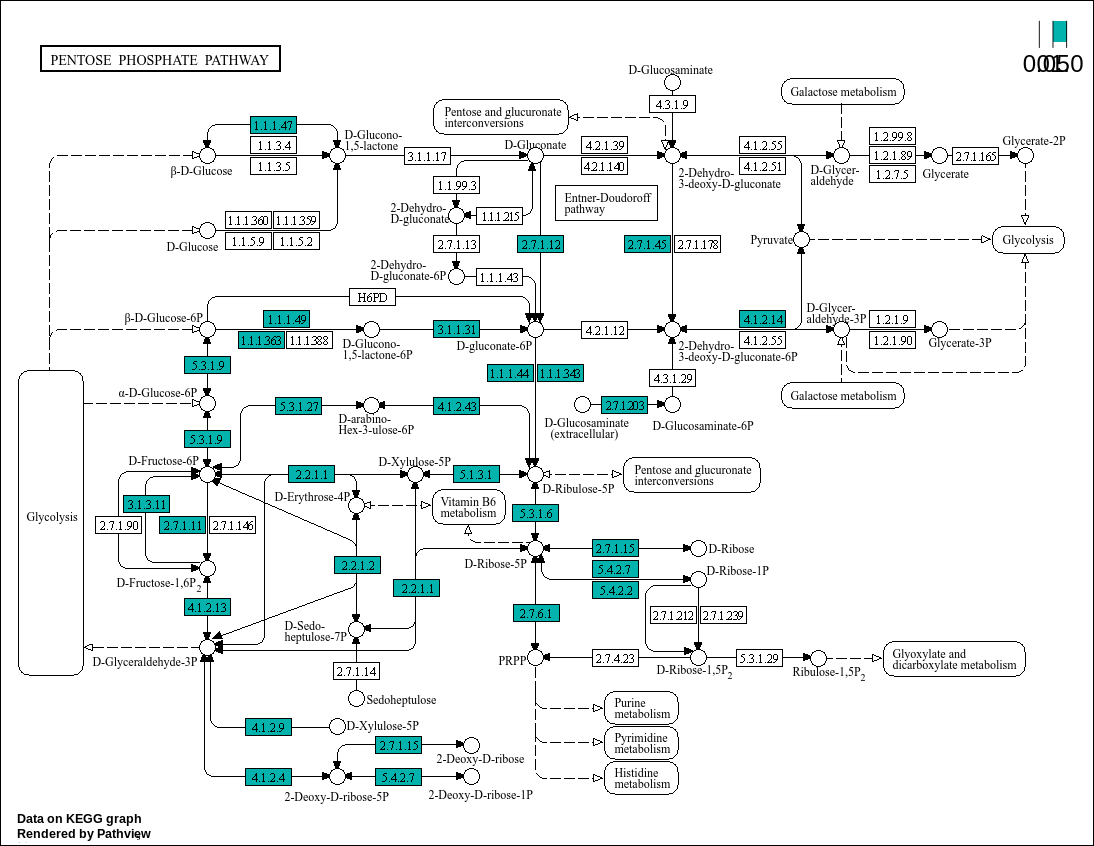
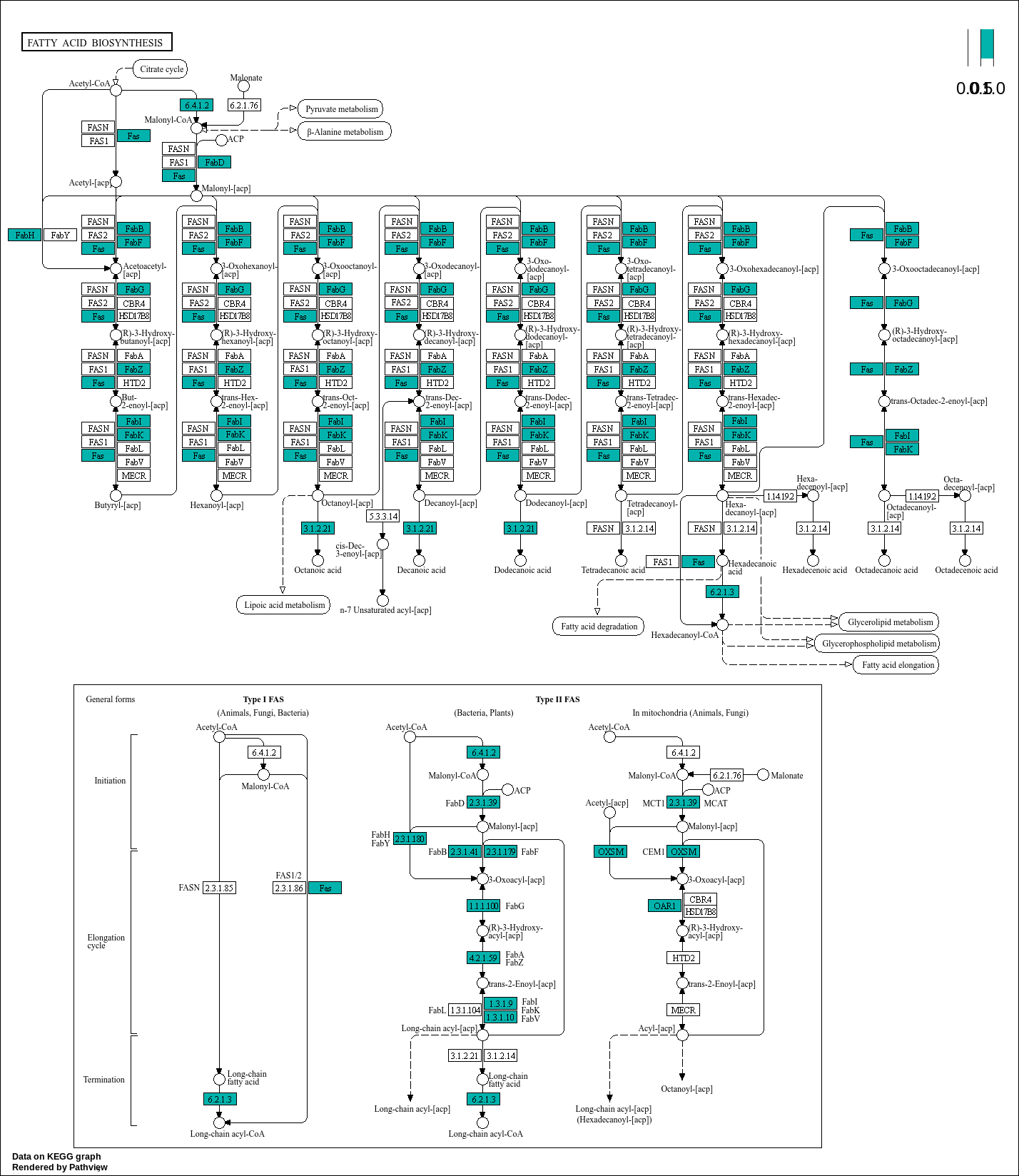
# Loading libraries:
suppressMessages({
library(ALDEx2)
library(data.table)
library(dplyr)
library(IRdisplay)
library(KEGGREST)
library(MGnifyR)
library(pathview)
library(tidyjson)
library(IRdisplay)
})
source("lib/variable_utils.r")
source("lib/kegg_pathways_utils.r")
display_markdown(file = '../_resources/mgnifyr_help.md')Help with MGnifyR
MGnifyR is an R package that provides a convenient way for R users to access data from the MGnify API.
Detailed help for each function is available in R using the standard ?function_name command (i.e. typing ?mgnify_query will bring up built-in help for the mgnify_query command).
A vignette is available containing a reasonably verbose overview of the main functionality. This can be read either within R with the vignette("MGnifyR") command, or in the development repository
MGnifyR Command cheat sheet
The following list of key functions should give a starting point for finding relevent documentation.
mgnify_client(): Create the client object required for all other functions.mgnify_query(): Search the whole MGnify database.mgnify_analyses_from_xxx(): Convert xxx accessions to analyses accessions. xxx is either samples or studies.mgnify_get_analyses_metadata(): Retrieve all study, sample and analysis metadata for given analyses.mgnify_get_analyses_phyloseq(): Convert abundance, taxonomic, and sample metadata into a single phyloseq object.mgnify_get_analyses_results(): Get functional annotation results for a set of analyses.mgnify_download(): Download raw results files from MGnify.mgnify_retrieve_json(): Low level API access helper function.